Home > Workload Solutions > High Performance Computing > White Papers > Dell Technologies Validated Design for Genomics with NVIDIA Clara Parabricks On AMD-powered Dell PowerEdge > Software performance evaluation
Software performance evaluation
-
Test case 1: Impact of NVLink
Clara Parabricks germline pipelines 3.7.0-1 and 3.6.1-1 are tested using PowerEdge XE8545 with four A100 GPUs (SMX4, NVLink version). The results with two A100 GPUs are compared from version 3.6.1-1 with PowerEdge R7525 for fair comparison. The main difference between the two server configurations is the number of GPUs and NVLink as shown in Table 3. Figure 4 shows the runtime difference with two A100s between two different servers with and without NVLink support. The runtime reductions between two servers with the Clara Parabricks version 3.6.1-1 are:
- 16% for 10x WGS data
- 32% for 30x WGS data
- 13% for 50x WGS data
There is no notable performance boost from version 3.6.1-1 to version 3.7.0-1 on the identical system. The Clara Parabricks team confirmed that version 3.7.0-1 is focused on the inclusion of new functionality and was not focused on additional accelerations.
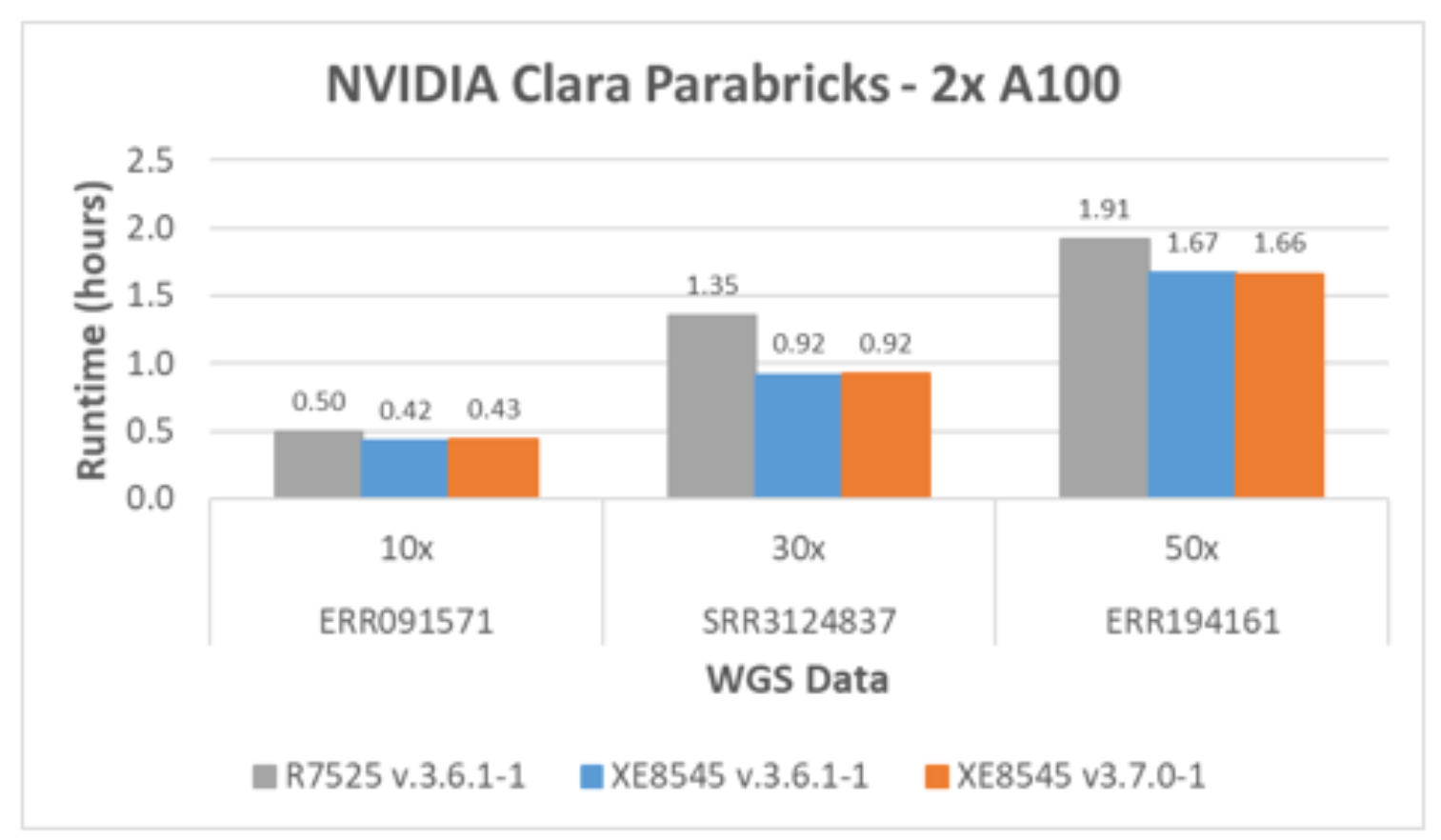
Figure 4. Parabricks germline pipeline performance with two A100 GPUs
Test Case 2: Continuous performance improvement through software development
NVIDIA continues to introduce software improvements to Clara Parabricks. The latest version, 3.7.0-1, focused on adding more tools than improving performance as shown in Figure 5. Dell Technologies has observed continuous performance improvement from an older version to a newer version.
Version 3.6.1 includes five somatic callers for comprehensive accelerated cancer genomic analysis: MuSE, LoFreq, Strelka2, Mutect2, and SomaticSnipter. In addition, tools were added to take advantage of archived data in scenarios when original FASTQ files were deleted to save storage space. BAM2FASTQ is an accelerated version of GATK Sam2fastq, which converts an existing BAM or CRAM file to a FASTQ file in version 3.7.0.
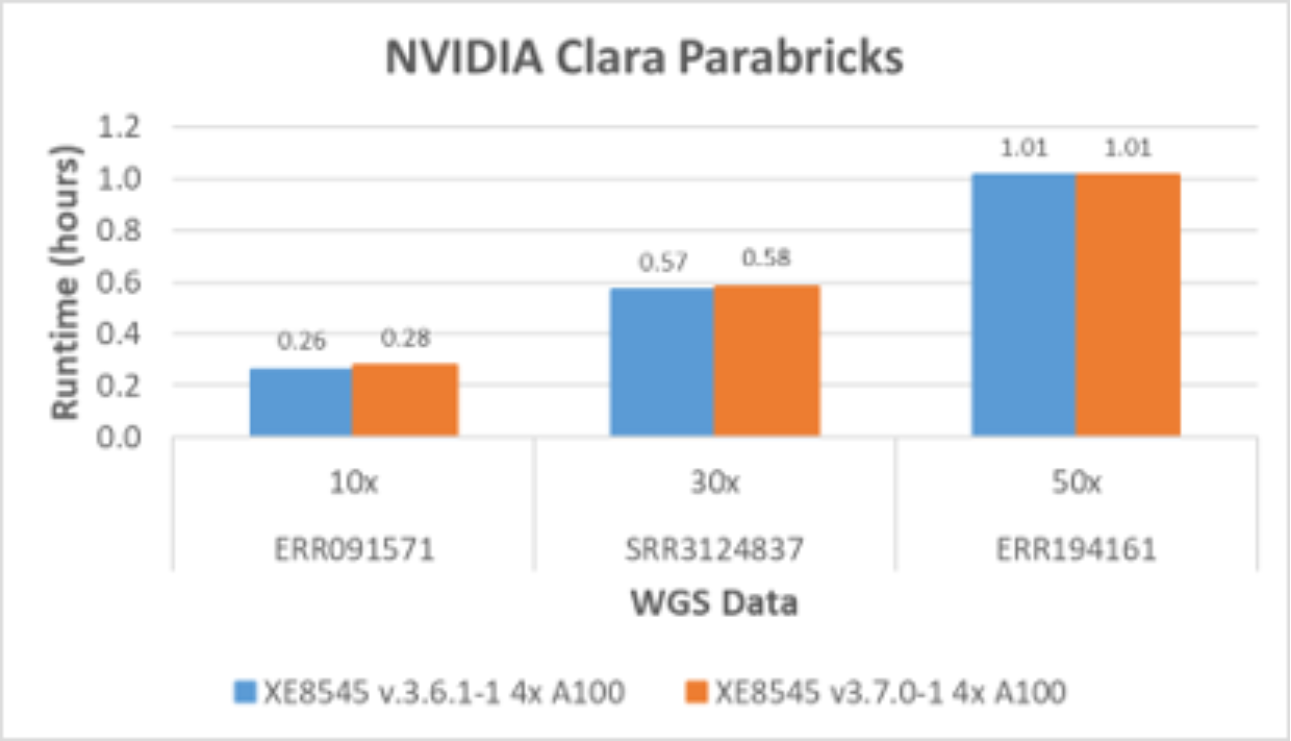
Figure 5. Parabricks germline pipeline performance with four A100 GPUs
Test Case 3: Scalability over data and GPUs
The minimum number of GPUs required for Clara Parabricks is two. As shown in Figure 6, the runtimes scale well for two and four GPUs with various sizes of WGS data. The previous test results with NVIDIA T4 GPUs show linear scalability up to 12 GPUs with 50x WGS data.
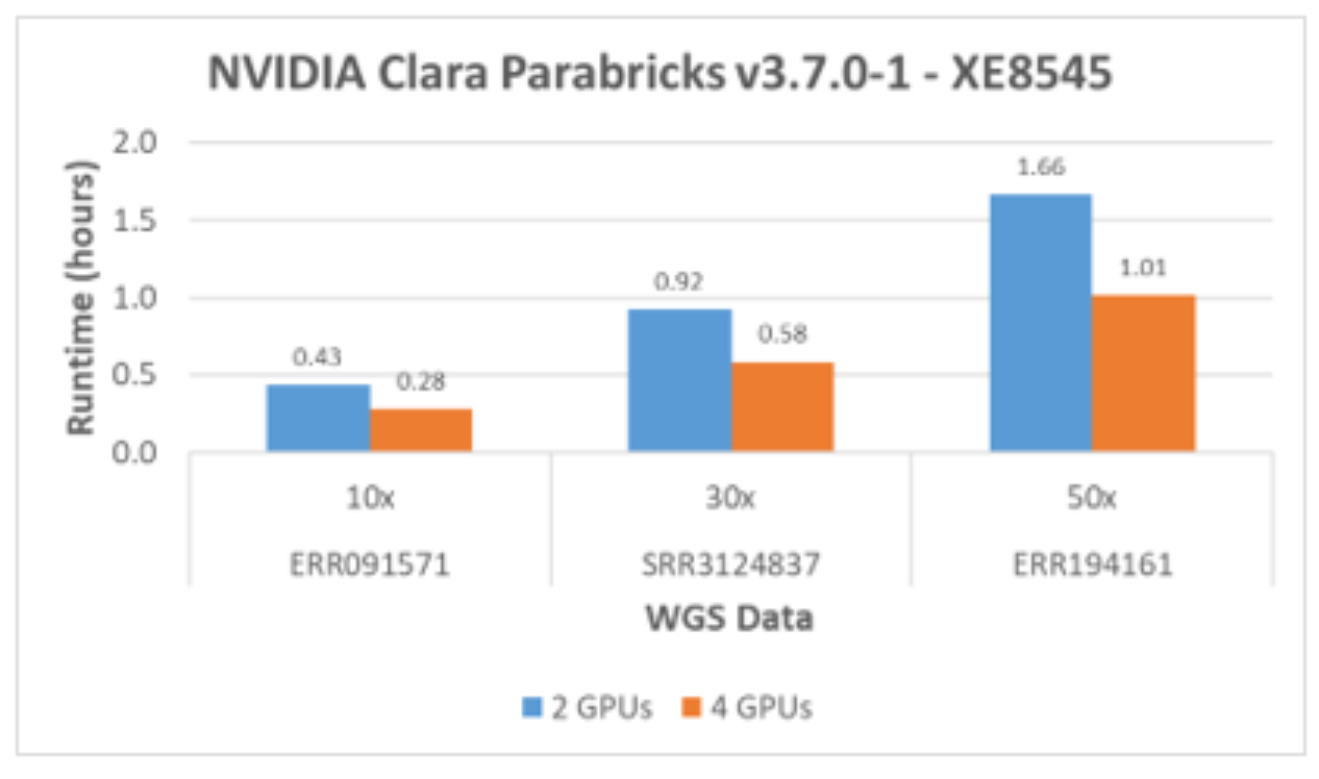
Figure 6. Scalability of A100 GPUs
