

Taming the Accelerator Cambrian Explosion with Omnia
Thu, 23 Sep 2021 18:29:00 -0000
|Read Time: 0 minutes
We are in the midst of a compute accelerator renaissance. Myriad new hardware accelerator companies are springing up with novel architectures and execution models for accelerating simulation and artificial intelligence (AI) workloads, each with a purported advantage over the others. Many are still in stealth, some have become public knowledge, others have started selling hardware, and still others have been gobbled up by larger, established players. This frenzied activity in the hardware space, driven by the growth of AI as a way to extract even greater value from new and existing data, has led some to liken it to the “Cambrian Explosion,” when life on Earth diversified at a rate not seen before or since.
If you’re in the business of standing up and maintaining infrastructure for high-performance computing and AI, this type of rapid diversification can be terrifying. How do I deal with all of these new hardware components? How do I manage all of the device drivers? What about all of the device plugins and operators necessary to make them function in my container-orchestrated environment? Data scientists and computational researchers often want the newest technology available, but putting it into production can be next to impossible. It’s enough to keep HPC/AI systems administrators lying awake at night.
At Dell Technologies, we now offer many different accelerator technologies within our PowerEdge server portfolio, from Graphics Processing Units (GPUs) in multiple sizes to Field-Programmable Gate Array (FPGA)-based accelerators. And there are even more to come. We understand that it can be a daunting task to manage all of this different hardware – it’s something we do every day in Dell Technologies’ HPC & AI Innovation Lab. So we’ve developed a mechanism for detecting, identifying, and deploying various accelerator technologies in an automated way, helping us to simplify our own deployment headaches. And we’ve integrated that capability into Omnia, an open-source, community-driven high-performance cluster deployment project started by Dell Technologies and Intel.
Deploy-time accelerator detection and installation
We recognize that tomorrow’s high-performance clusters will not be fully homogenous, consisting of exact copies of the same compute building block replicated tens, hundreds, or thousands of times. Instead clusters are becoming more heterogeneous, consisting of as many as a dozen different server configurations, all tied together under a single (or in some cases – multiple) scheduler or container orchestrator.
This heterogeneity can be a problem for many of today’s cluster deployment tools, which rely on the concept of the “golden image” – a complete image of the server's operating system, hardware drivers, and software stack. The golden image model is extremely useful in many environments, such as homogeneous and diskless deployments. But in the clusters of tomorrow, which will try to capture the amazing potential of this hardware diversity, the golden image model becomes unmanageable.
Instead, Omnia does not rely on the golden image. We think of cluster deployment like 3D-printing – rapidly placing layer after layer of software components and capabilities on top of the hardware until a functional server building block emerges. This allows us, with the use of some intelligent detection and logic, to build bespoke software stacks for each server building block; on demand, at deploy time. From Omnia’s perspective, there’s really no difference between deploying a compute server with no accelerators into a cluster versus deploying a compute server with GPUs or FPGAs into that same cluster. We simply pick different component layers during the process.
What does this mean for cluster deployment?
It means that clusters can now be built from a variety of heterogeneous server building blocks, all managed together as a single entity. Instead of a cluster of CPU servers, another cluster of GPU-accelerated servers, and yet another cluster of FPGA-accelerated servers, research and HPC IT organizations can manage a single resource with all of the different types of technologies that their users demand, all connected by a unified network fabric and sharing a set of unified storage solutions.
And by using Omnia, the process of deploying clusters of heterogeneous building blocks has been dramatically simplified. Regardless of how many types of building blocks an organization wants to use within their next-generation cluster, it can all be deployed using the same approach, and at the same time. There’s no need to build special images for this type of server and that type of server, simply start the Omnia deployment process and Omnia’s intelligent software deployment system will do the rest.
Learn more
Omnia is available to download on GitHub today. You can learn more about the Omnia project in our previous blog post.
Related Blog Posts

Omnia: Open-source deployment of high-performance clusters to run simulation, AI, and data analytics workloads
Mon, 12 Dec 2022 18:31:28 -0000
|Read Time: 0 minutes
High Performance Computing (HPC), in which clusters of machines work together as one supercomputer, is changing the way we live and how we work. These clusters of CPU, memory, accelerators, and other resources help us forecast the weather and understand climate change, understand diseases, design new drugs and therapies, develop safe cars and planes, improve solar panels, and even simulate life and the evolution of the universe itself. The cluster architecture model that makes this compute-intensive research possible is also well suited for high performance data analytics (HPDA) and developing machine learning models. With the Big Data era in full swing and the Artificial Intelligence (AI) gold rush underway, we have seen marketing teams with their own Hadoop clusters attempting to transition to HPDA and finance teams managing their own GPU farms. Everyone has the same goals: to gain new, better insights faster by using HPDA and by developing advanced machine learning models using techniques such as deep learning and reinforcement learning. Today, everyone has a use for their own high-performance computing cluster. It’s the age of the clusters!
Today's AI-driven IT Headache: Siloed Clusters and Cluster Sprawl
Unfortunately, cluster sprawl has taken over our data centers and consumes inordinate amounts of IT resources. Large research organizations and businesses have a cluster for this and a cluster for that. Perhaps each group has a little “sandbox” cluster, or each type of workload has a different cluster. Many of these clusters look remarkably similar, but they each need a dedicated system administrator (or set of administrators), have different authorization credentials, different operating models, and sit in different racks in your data center. What if there was a way to bring them all together?
That’s why Dell Technologies, in partnership with Intel, started the Omnia project.
The Omnia Project
The Omnia project is an open-source initiative with a simple aim: To make consolidated infrastructure easy and painless to deploy using open open source and free use software. By bringing the best open source software tools together with the domain expertise of Dell Technologies' HPC & AI Innovation Lab, HPC & AI Centers of Excellence, and the broader HPC Community, Omnia gives customers decades of accumulated expertise in deploying state-of-the-art systems for HPC, AI, and Data Analytics – all in a set of easily executable Ansible playbooks. In a single day, a stack of servers, networking switches, and storage arrays can be transformed into one consolidated cluster for running all your HPC, AI, and Data Analytics workloads. Omnia project logo
Omnia project logo
Simple by Design
Omnia’s design philosophy is simplicity. We look for the best, most straightforward approach to solving each task.
- Need to run the Slurm workload manager? Omnia assembles Ansible plays which build the right rpm files and deploy them correctly, making sure all the correct dependencies are installed and functional.
- Need to run the Kubernetes container orchestrator? Omnia takes advantage of community supported package repositories for Linux (currently CentOS) and automates all the steps for creating a functional multi-node Kubernetes cluster.
- Need a multi-user, interactive Python/R/Julia development environment? Omnia takes advantage of best-of-breed deployments for Kubernetes through Helm and OperatorHub, provides configuration files for dynamic and persistent storage, points to optimized containers in DockerHub, Nvidia GPU Cloud (NGC), or other container registries for unaccelerated and accelerated workloads, and automatically deploys machine learning platforms like Kubeflow.
Before we go through the process of building something from scratch, we will make sure there isn’t already a community actively maintaining that toolset. We’d rather leverage others' great work than reinvent the wheel.
Inclusive by Nature
Omnia’s contribution philosophy is inclusivity. From code and documentation updates to feature requests and bug reports, every user’s contributions are welcomed with open arms. We provide an open forum for conversations about feature ideas and potential implementation solutions, making use of issue threads on GitHub. And as the project grows and expands, we expect the technical governance committee to grow to include the top contributors and stakeholders from the community.
What's Next?
Omnia is just getting started. Right now, we can easily deploy Slurm and Kubernetes clusters from a stack of pre-provisioned, pre-networked servers, but our aim is higher than that. We are currently adding capabilities for performing bare-metal provisioning and supporting new and varying types of accelerators. In the future, we want to collect information from the iDRAC out-of-band management system on Dell EMC PowerEdge servers, configure Dell EMC PowerSwitch Ethernet switches, and much more.
What does the future hold? While we have plans in the near-term for additional feature integrations, we are looking to partner with the community to define and develop future integrations. Omnia will grow and develop based on community feedback and your contributions. In the end, the Omnia project will not only install and configure the open source software we at Dell Technologies think is important, but the software you – the community – want it to, as well! We can’t think of a better way for our customers to be able to easily setup clusters for HPC, AI, and HPDA workloads, all while leveraging the expertise of the entire Dell Technologies' HPC Community.
Omnia is available today on GitHub at https://github.com/dellhpc/omnia. Join the community now and help guide the design and development of the next generation of open-source consolidated cluster deployment tools!

Training an AI Radiologist with Distributed Deep Learning
Wed, 07 Dec 2022 13:25:02 -0000
|Read Time: 0 minutes
Originally published on Aug 16, 2018 11:14:00 AM
The potential of neural networks to transform healthcare is evident. From image classification to dictation and translation, neural networks are achieving or exceeding human capabilities. And they are only getting better at these tasks as the quantity of data increases.
But there’s another way in which neural networks can potentially transform the healthcare industry: Knowledge can be replicated at virtually no cost. Take radiology as an example: To train 100 radiologists, you must teach each individual person the skills necessary to identify diseases in x-ray images of patients’ bodies. To make 100 AI-enabled radiologist assistants, you take the neural network model you trained to read x-ray images and load it into 100 different devices.
The hurdle is training the model. It takes a large amount of cleaned, curated, labeled data to train an image classification model. Once you’ve prepared the training data, it can take days, weeks, or even months to train a neural network. Even once you’ve trained a neural network model, it might not be smart enough to perform the desired task. So, you try again. And again. Eventually, you will train a model that passes the test and can be used out in the world.
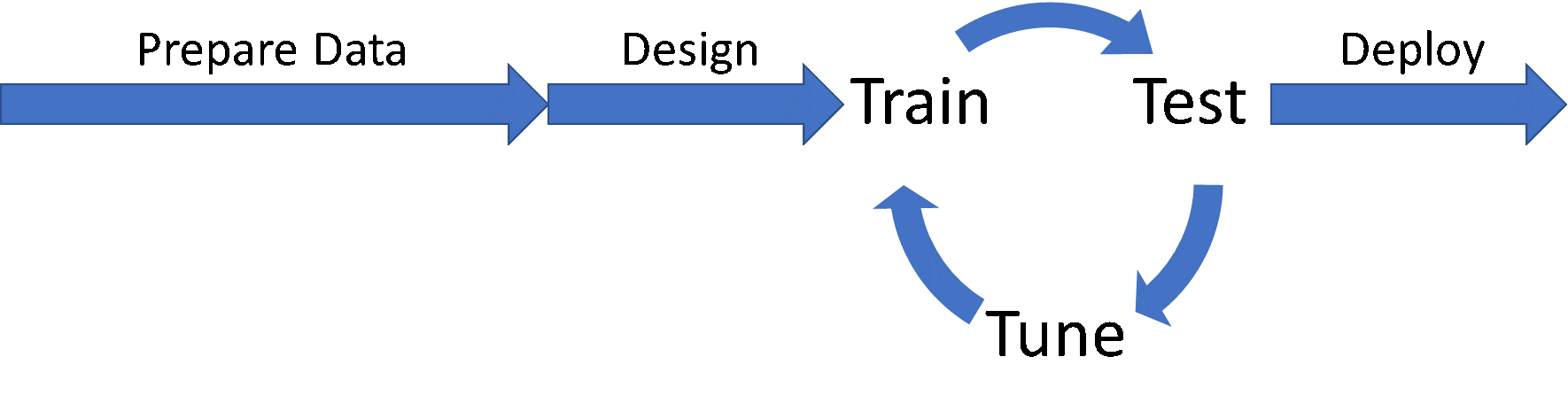 Workflow for developing neural network modelsIn this post, I’m going to talk about how to reduce the time spent in the Train/Test/Tune cycle by speeding up the training portion with distributed deep learning, using a test case we developed in Dell EMC’s HPC & AI Innovation Lab to classify pathologies in chest x-ray images. Through a combination of distributed deep learning, optimizer selection, and neural network topology selection, we were able to not only speed the process of training models from days to minutes, we were also able to improve the classification accuracy significantly.
Workflow for developing neural network modelsIn this post, I’m going to talk about how to reduce the time spent in the Train/Test/Tune cycle by speeding up the training portion with distributed deep learning, using a test case we developed in Dell EMC’s HPC & AI Innovation Lab to classify pathologies in chest x-ray images. Through a combination of distributed deep learning, optimizer selection, and neural network topology selection, we were able to not only speed the process of training models from days to minutes, we were also able to improve the classification accuracy significantly.
Starting Point: Stanford University’s CheXNet
We began by surveying the landscape of AI projects in healthcare, and Andrew Ng’s group at Stanford University provided our starting point. CheXNet was a project to demonstrate a neural network’s ability to accurately classify cases of pneumonia in chest x-ray images.
The dataset that Stanford used was ChestXray14, which was developed and made available by the United States’ National Institutes of Health (NIH). The dataset contains over 120,000 images of frontal chest x-rays, each potentially labeled with one or more of fourteen different thoracic pathologies. The data set is very unbalanced, with more than half of the data set images having no listed pathologies.
Stanford decided to use DenseNet, a neural network topology which had just been announced as the Best Paper at the 2017 Conference on Computer Vision and Pattern Recognition (CVPR), to solve the problem. The DenseNet topology is a deep network of repeating blocks over convolutions linked with residual connections. Blocks end with a batch normalization, followed by some additional convolution and pooling to link the blocks. At the end of the network, a fully connected layer is used to perform the classification.
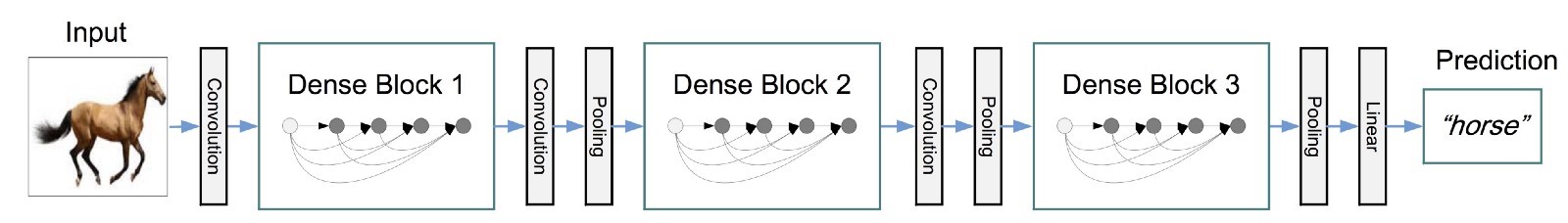 An illustration of the DenseNet topology (source: Kaggle)
An illustration of the DenseNet topology (source: Kaggle)
Stanford’s team used a DenseNet topology with the layer weights pretrained on ImageNet and replaced the original ImageNet classification layer with a new fully connected layer of 14 neurons, one for each pathology in the ChestXray14 dataset.
Building CheXNet in Keras
It’s sounds like it would be difficult to setup. Thankfully, Keras (provided with TensorFlow) provides a simple, straightforward way of taking standard neural network topologies and bolting-on new classification layers.
from tensorflow import keras from keras.applications import DenseNet121 orig_net = DenseNet121(include_top=False, weights='imagenet', input_shape=(256,256,3))
In this code snippet, we are importing the original DenseNet neural network (DenseNet121) and removing the classification layer with the include_top=False argument. We also automatically import the pretrained ImageNet weights and set the image size to 256x256, with 3 channels (red, green, blue).
With the original network imported, we can begin to construct the classification layer. If you look at the illustration of DenseNet above, you will notice that the classification layer is preceded by a pooling layer. We can add this pooling layer back to the new network with a single Keras function call, and we can call the resulting topology the neural network's filters, or the part of the neural network which extracts all the key features used for classification.
from keras.layers import GlobalAveragePooling2D filters = GlobalAveragePooling2D()(orig_net.output)
The next task is to define the classification layer. The ChestXray14 dataset has 14 labeled pathologies, so we have one neuron for each label. We also activate each neuron with the sigmoid activation function, and use the output of the feature filter portion of our network as the input to the classifiers.
from keras.layers import Dense classifiers = Dense(14, activation='sigmoid', bias_initializer='ones')(filters)
The choice of sigmoid as an activation function is due to the multi-label nature of the data set. For problems where only one label ever applies to a given image (e.g., dog, cat, sandwich), a softmax activation would be preferable. In the case of ChestXray14, images can show signs of multiple pathologies, and the model should rightfully identify high probabilities for multiple classifications when appropriate.
Finally, we can put the feature filters and the classifiers together to create a single, trainable model.
from keras.models import Model chexnet = Model(inputs=orig_net.inputs, outputs=classifiers)
With the final model configuration in place, the model can then be compiled and trained.
Accelerating the Train/Test/Tune Cycle with Distributed Deep Learning
To produce better models sooner, we need to accelerate the Train/Test/Tune cycle. Because testing and tuning are mostly sequential, training is the best place to look for potential optimization.
How exactly do we speed up the training process? In Accelerating Insights with Distributed Deep Learning, Michael Bennett and I discuss the three ways in which deep learning can be accelerated by distributing work and parallelizing the process:
- Parameter server models such as in Caffe or distributed TensorFlow,
- Ring-AllReduce approaches such as Uber’s Horovod, and
- Hybrid approaches for Hadoop/Spark environments such as Intel BigDL.
Which approach you pick depends on your deep learning framework of choice and the compute environment that you will be using. For the tests described here we performed the training in house on the Zenith supercomputer in the Dell EMC HPC & AI Innovation Lab. The ring-allreduce approach enabled by Uber’s Horovod framework made the most sense for taking advantage of a system tuned for HPC workloads, and which takes advantage of Intel Omni-Path (OPA) networking for fast inter-node communication. The ring-allreduce approach would also be appropriate for solutions such as the Dell EMC Ready Solutions for AI, Deep Learning with NVIDIA.
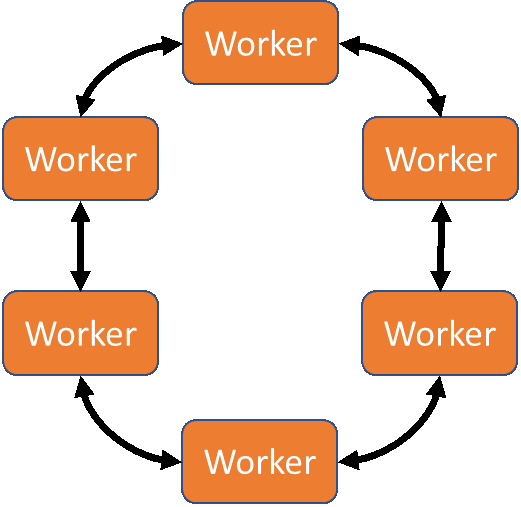 The MPI-RingAllreduce approach to distributed deep learning
The MPI-RingAllreduce approach to distributed deep learning
Horovod is an MPI-based framework for performing reduction operations between identical copies of the otherwise sequential training script. Because it is MPI-based, you will need to be sure that an MPI compiler (mpicc) is available in the working environment before installing horovod.
Adding Horovod to a Keras-defined Model
Adding Horovod to any Keras-defined neural network model only requires a few code modifications:
- Initializing the MPI environment,
- Broadcasting initial random weights or checkpoint weights to all workers,
- Wrapping the optimizer function to enable multi-node gradient summation,
- Average metrics among workers, and
- Limiting checkpoint writing to a single worker.
Horovod also provides helper functions and callbacks for optional capabilities that are useful when performing distributed deep learning, such as learning-rate warmup/decay and metric averaging.
Initializing the MPI Environment
Initializing the MPI environment in Horovod only requires calling the init method:
import horovod.keras as hvd hvd.init()
This will ensure that the MPI_Init function is called, setting up the communications structure and assigning ranks to all workers.
Broadcasting Weights
Broadcasting the neuron weights is done using a callback to the Model.fit Keras method. In fact, many of Horovod’s features are implemented as callbacks to Model.fit, so it’s worthwhile to define a callback list object for holding all the callbacks.
callbacks = [ hvd.callbacks.BroadcastGlobalVariablesCallback(0) ]
You’ll notice that the BroadcastGlobalVariablesCallback takes a single argument that’s been set to 0. This is the root worker, which will be responsible for reading checkpoint files or generating new initial weights, broadcasting weights at the beginning of the training run, and writing checkpoint files periodically so that work is not lost if a training job fails or terminates.
Wrapping the Optimizer Function
The optimizer function must be wrapped so that it can aggregate error information from all workers before executing. Horovod’s DistributedOptimizer function can wrap any optimizer which inherits Keras’ base Optimizer class, including SGD, Adam, Adadelta, Adagrad, and others.
import keras.optimizers opt = hvd.DistributedOptimizer(keras.optimizers.Adadelta(lr=1.0))
The distributed optimizer will now use the MPI_Allgather collective to aggregate error information from training batches onto all workers, rather than collecting them only to the root worker. This allows the workers to independently update their models rather than waiting for the root to re-broadcast updated weights before beginning the next training batch.
Averaging Metrics
Between steps error metrics need to be averaged to calculate global loss. Horovod provides another callback function to do this called MetricAverageCallback.
callbacks = [ hvd.callbacks.BroadcastGlobalVariablesCallback(0), hvd.callbacks.MetricAverageCallback() ]
This will ensure that optimizations are performed on the global metrics, not the metrics local to each worker.
Writing Checkpoints from a Single Worker
When using distributed deep learning, it’s important that only one worker write checkpoint files to ensure that multiple workers writing to the same file does not produce a race condition, which could lead to checkpoint corruption.
Checkpoint writing in Keras is enabled by another callback to Model.fit. However, we only want to call this callback from one worker instead of all workers. By convention, we use worker 0 for this task, but technically we could use any worker for this task. The one good thing about worker 0 is that even if you decide to run your distributed deep learning job with only 1 worker, that worker will be worker 0.
callbacks = [ ... ]
if hvd.rank() == 0:
callbacks.append(keras.callbacks.ModelCheckpoint('./checkpoint-{epoch].h5'))Result: A Smarter Model, Faster!
Once a neural network can be trained in a distributed fashion across multiple workers, the Train/Test/Tune cycle can be sped up dramatically.
The figure below shows exactly how dramatically. The three tests shown are the training speed of the Keras DenseNet model on a single Zenith node without distributed deep learning (far left), the Keras DenseNet model with distributed deep learning on 32 Zenith nodes (64 MPI processes, 2 MPI processes per node, center), and a Keras VGG16 version using distributed deep learning on 64 Zenith nodes (128 MPI processes, 2 MPI processes per node, far right). By using 32 nodes instead of a single node, distributed deep learning was able to provide a 47x improvement in training speed, taking the training time for 10 epochs on the ChestXray14 data set from 2 days (50 hours) to less than 2 hours!
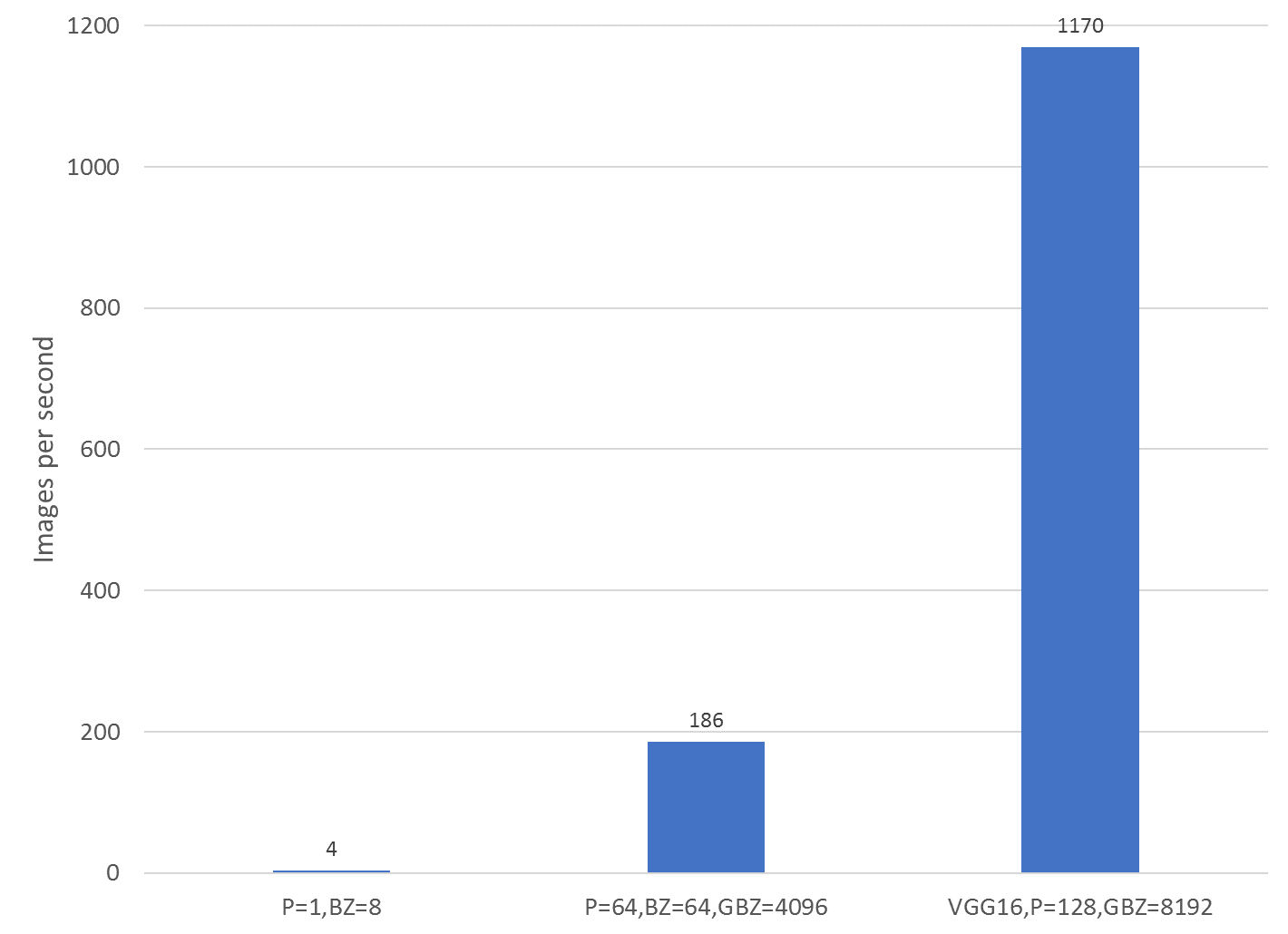 Performance comparisons of Keras models with distributed deep learning using Horovod
Performance comparisons of Keras models with distributed deep learning using Horovod
The VGG variant, trained on 128 Zenith nodes, was able to complete the same number of epochs as was required for the single-node DenseNet version to train in less than an hour, although it required more epochs to train. It also, however, was able to converge to a higher-quality solution. This VGG-based model outperformed the baseline, single-node model in 4 of 14 conditions, and was able to achieve nearly 90% accuracy in classifying emphysema.
 Accuracy comparison of baseline single-node DenseNet model vs VGG variant with Horovod
Accuracy comparison of baseline single-node DenseNet model vs VGG variant with Horovod
Conclusion
In this post we’ve shown you how to accelerate the Train/Test/Tune cycle when developing neural network-based models by speeding up the training phase with distributed deep learning. We walked through the process of transforming a Keras-based model to take advantage of multiple nodes using the Horovod framework, and how these few simple code changes, coupled with some additional compute infrastructure, can reduce the time needed to train a model from days to minutes, allowing more time for the testing and tuning pieces of the cycle. More time for tuning means higher-quality models, which means better outcomes for patients, customers, or whomever will benefit from the deployment of your model.
Lucas A. Wilson, Ph.D. is the Chief Data Scientist in Dell EMC's HPC & AI Innovation Lab. (Twitter: @lucasawilson)



