

Scaling Hardware and Computation for Practical Deep Learning Applications in Healthcare
Fri, 01 Dec 2023 15:26:29 -0000
|Read Time: 0 minutes
Medical practice requires analysis of large volumes of data spanning multiple modalities. While these can be as simple as numeric lab results, at the other extreme are high-complexity data such as magnetic resonance imaging or decades-worth of text-based clinical documentation which may be present in medical records. Oftentimes, small details buried within piles of clinical information are critical to obtaining a complete clinical picture. Many deep learning methods developed in recent years have focused on very short “sequence lengths” – the term used to describe the number of words or pixels that a model can ingest – of images and text compared to those encountered in clinical practice. How do we scale such tools to model this breadth of clinical data appropriately and efficiently?
In the following blog, we discuss ways to tackle the compute requirements of developing transformer-based deep learning tools for healthcare data from the hardware, data processing, and modeling perspectives. To do so, we present a practical application of Flash Attention using a series of experiments performing an analysis of the publicly available Kaggle RSNA Screening Mammography Breast Cancer Detection challenge, which contains 54,706 images of 11,913 patients. Breast cancer affects 1 in 8 women and is the second leading cause of cancer death. As such, screening mammography is one of the most performed imaging-based medical screening procedures, which offers a clinically relevant and data-centric case study to consider.
Data Primer
To detect breast cancer early when treatments are most effective, high-resolution x-ray images are taken of breast tissue to identify areas of abnormality which require further examination by biopsy or more detailed imaging. Typically, two views are acquired:
- Craniocaudal (CC) – taken from the head-to-toe perspective
- Mediolateral oblique (MLO) – taken at an angle
The dataset contains DICOM-formatted images which must be pre-processed in a standard fashion prior to model training. We detail the data preparation pipeline in figure 1. The CC and MLO views of each study are identified, flipped horizontally if necessary, cropped, and combined to form the model input image. We wrap the standard PyTorch Dataset class to load images and preprocess them for training.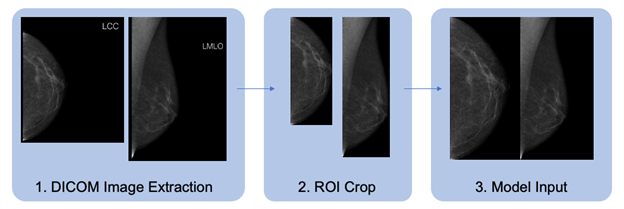 Figure 1. Data pre-processing pipeline for DICOM-formatted image
Figure 1. Data pre-processing pipeline for DICOM-formatted image
A more in-depth look at the system for data pre-processing is as follows:
- For each breast with a corresponding cancer label, the CC and MLO views are extracted, and the image data are normalized. Right-sided images are horizontally flipped so that the tissue is to the left side of the image, as shown.
- Images are cropped to the region of interest (ROI), excluding areas of black or non-tissue artifacts.
- Images are resized, maintaining aspect ratio, and tiled to a square of the output size of interest, with the CC view occupying the left half of the output and the MLO view occupying the right.
An important consideration is whether to perform this processing within the dataloader while training or to save a pre-processed version of the dataset. The former approach allows for iteration on different processing strategies without modifying the dataset itself, providing greater ease of experimentation. However, this level of processing during training may limit the rate at which data can be fed to the graphics processing unit (GPU) for training, resulting in time and monetary inefficiencies. In contrast, the latter approach requires that multiple versions of the dataset be saved locally, which is potentially prohibitive when working with large dataset sizes and storage space and/or network limitations. For the purpose of this blog post, to benchmark GPU hardware and training optimizations, we use the second method, saving data on local solid state drives connected via NVMe to ensure GPU saturation despite processor differences. In general, before implementing the training optimizations described below, it is important to first ensure that dataloading does not bottleneck the overall training process.
Scaling Up
Naturally, increasing the capability and amount of compute available for model training yields direct benefits. To demonstrate the influence of hardware on run time, we present a simple 20-epoch training experiment using the same dataset on three different servers, shown in figure 2:
- Dell XE8545 with 4x NVIDIA A100-SXM4 40GB GPUs and an AMD EPYC 7763 with 64 cores
- Dell R750xa with 4x NVIDIA A100 80GB GPUs and an Intel Xeon Gold 5320 processor with 26 cores
- Dell XE9680 server with 8 NVIDIA HGX A100 80GB SXM4 GPUs and an Intel Xeon Platinum 8470 processor with 52 cores
Input data into the model shown in figure 2 were 512x512 with a patch size of 16. Batch size was 24 per GPU on the 40GB and 64 on the 80GB servers.
Parameters remain the same for each run, except that batch size has been increased to maximally utilize GPU memory on the R750xa and XE9680 compared with the XE8545. Gradient accumulation is performed to maintain a constant global batch size per model weight update for each run. We see a clear improvement in runtime as the hardware is scaled up, demonstrating how increased compute capability directly yields time savings which enables researchers to efficiently iterate on experiments and train effective models.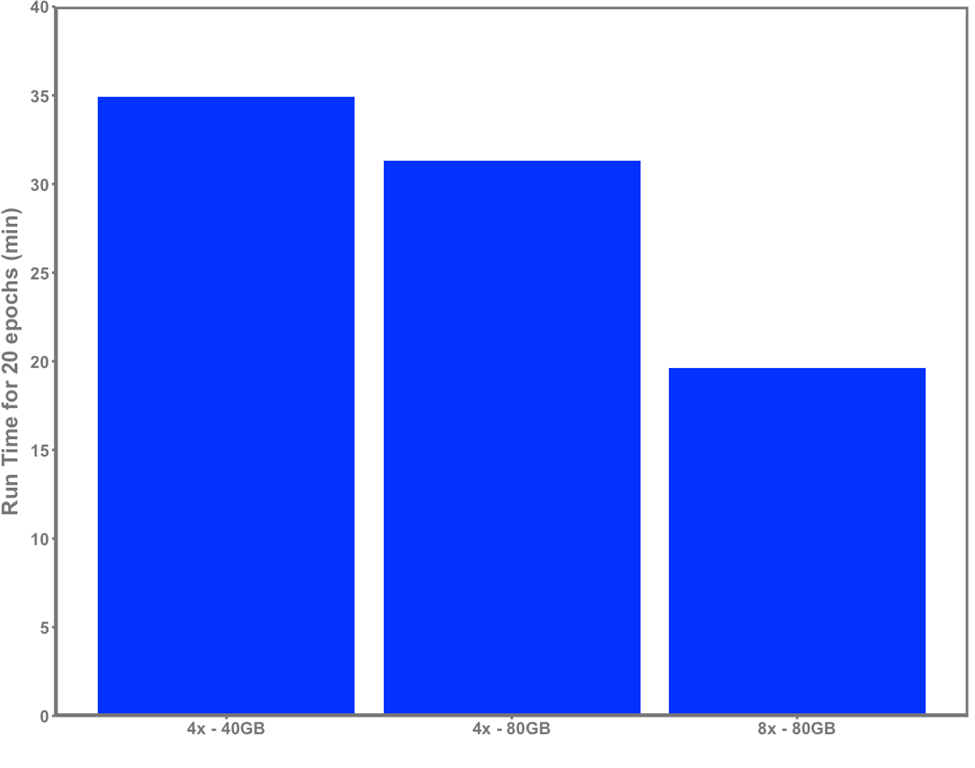 Figure 2. ViT-base training time across 20 epochs with 4xA100 40GB, 4xA100 80GB, and 8xA100 80GB servers
Figure 2. ViT-base training time across 20 epochs with 4xA100 40GB, 4xA100 80GB, and 8xA100 80GB servers
In conjunction with hardware, sequence lengths of data should be carefully considered given the application of interest. The selected tokenization scheme directly impacts sequence length of input data, such as the patch size selected as input to a vision transformer. For example, a patch size of 16 on a 1024x1024 image will result in a sequence length of 4,096 (Height*Width/Patch Size2) while a patch size of 8 will result in a sequence length of 16,384. While GPUs increasingly feature more memory, they present an upper bound on the sequence length that can practicably be considered. Smaller patch sizes – and thus, longer sequences – will result in slower throughput via smaller batch sizes and a greater number of computations, as shown in figure 3. However, larger images sizes coupled with smaller patch sizes are particularly relevant in analysis of mammography and other applications in which fine-resolution features are of interest.
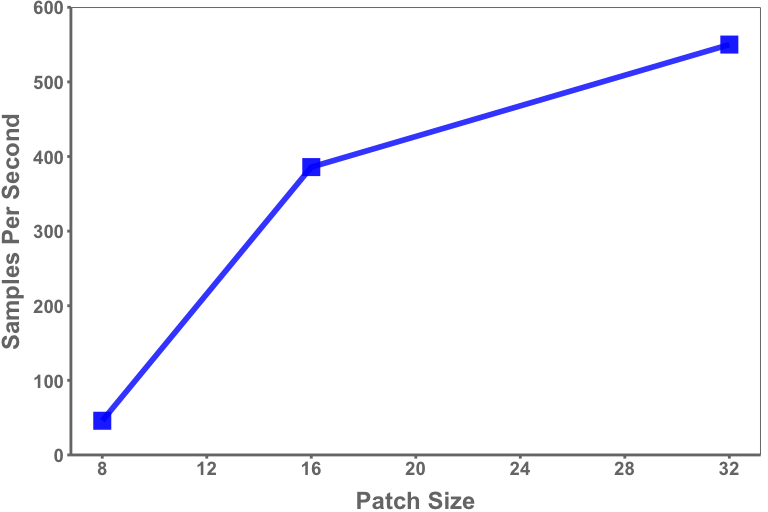 Figure 3. Average training samples per second (per GPU) for mammograms through a vision transformer by patch size
Figure 3. Average training samples per second (per GPU) for mammograms through a vision transformer by patch size
The data illustrated in figure 3 are taken from a run of twenty epochs using an image size of 512x512 and tested on an 8xA100 (80 GB) server.
Flash Attention – Experiments
Recently, Dao et al. have published on Flash Attention (https://arxiv.org/abs/2205.14135), a technique aimed at more efficiently accomplishing the computations involved within transformers via minimizing GPU high-bandwidth memory and the on-chip SRAM. Their reported findings are impressive, yielding 2-3x speedups during an attention forward and backwards pass while also having 3-20x smaller memory requirements.
Using a Dell XE9680 server with 8 NVIDIA HGX A100 80GB SXM4 GPUs and an Intel Xeon Platinum 8470 processor with 52 cores, we provide a practical demonstration of potential applications for Flash Attention and vision transformers in healthcare. Specifically, we performed experiments to demonstrate how sequence length (determined by patch size and image size) and Flash Attention impact training time. To limit confounding variables, all images were pre-sized on disk and directly loaded into the vision transformer without any permutations. For the vision transformer, the ViT-Base from Huggingface was used. For Flash Attention, the Encoder from the x_transformers library was used, shown being implemented in the following code.
All tests were carried out with the Huggingface trainer using an effective batch size of 128 per GPU, “brain" floating-point 16 data, and across twenty epochs at patch sizes of 8, 16, and 32 with image sizes of 384, 512, 1024, and 2048.
from x_transformers import ViTransformerWrapper, Encoder
class FlashViT(nn.Module):
def __init__(self,
encoder = ViTransformerWrapper(
image_size = args.img_size,
patch_size = args.patch_size,
num_classes = 2,
channels=3,
attn_layers = Encoder(
dim = 768,
depth = 12,
heads = 12,
attn_flash=True
)
),
super().__init__()
self.encoder = encoder
def forward(self,
pixel_values:torch.tensor,
labels:torch.tensor):
"""
pixel_values: [batch,channel,ht,wt] of pixel values
labels: labels for each image
"""
logits = self.encoder(pixel_values)
return {'loss':F.cross_entropy(logits,labels),'logits':logits}
model = FlashViT()
Figure 4 demonstrates the pronounced benefit of using Flash Attention within a vision transformer with respect to model throughput. With the exception of the two smallest image sizes and largest patch size (and thus shortest sequence length), Flash Attention resulted in a marked speed increase across all other perturbations. The speed-up range across patch sizes was:
- Patch size of 8: 3.0 - 4.2x
- Patch size of 16: 2.8 – 4.0x
- Patch size of 32: 0 - 2.3x
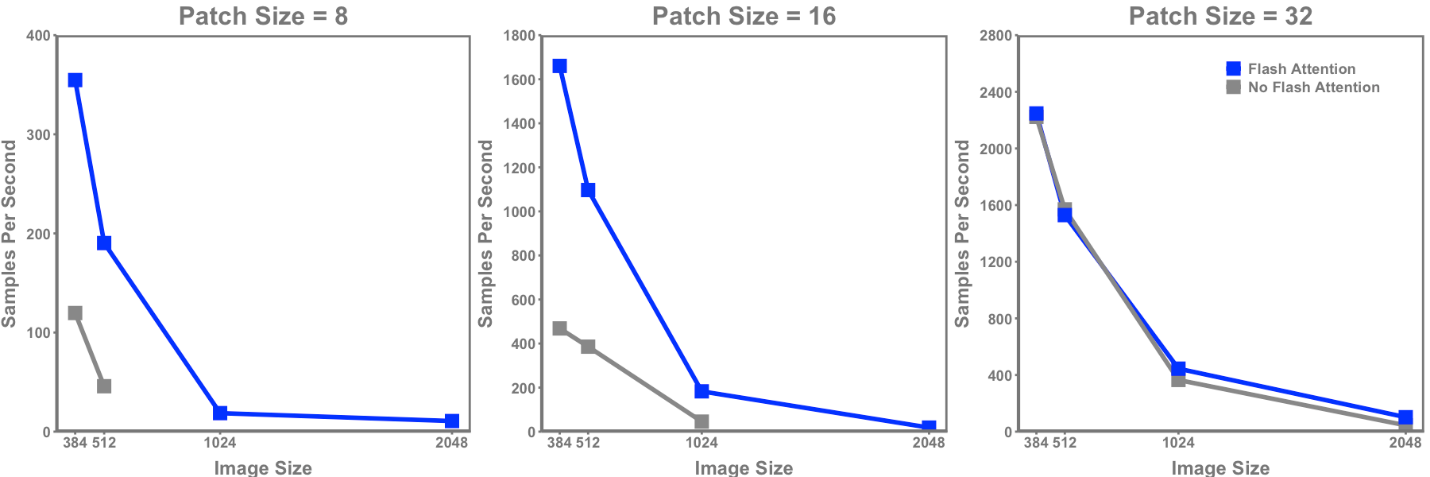 Figure 4. Samples per second throughput for a ViT-base vision transformer with and without Flash Attention across varying image sizes and patch sizes
Figure 4. Samples per second throughput for a ViT-base vision transformer with and without Flash Attention across varying image sizes and patch sizes
Another benefit demonstrated in these experiments is the additional image and patch size combinations achievable only with Flash Attention due to the reduced GPU memory requirement. Non-Flash Attention models could only be used on image sizes of 2,048 if a patch size of 32 was used (sequence length of 4,096), whereas Flash Attention was capable of running on patch sizes of 8 and 16. Even at shorter sequence lengths (576 - 384x384 image, patch size of 16), there was 2.3x less memory used for Flash Attention. Use of Flash Attention will also be critical when considering larger transformer models, with ViT-Huge having more than 7x the parameters than ViT-Base. In conjunction with hardware-enabling distributed training at scale such as the Dell XE9680, these optimizations will enable new findings at unprecedented scales.
Takeaways
We have described methods by which the benefits of transformer-based models can be scaled to the longer sequences which medical data often require. Notably, we demonstrate the benefits of implementing Flash Attention to a vision encoder. Flash Attention presents marked benefit from a modeling perspective, from shorter runtimes (and thus lower cost) to better image encoding (longer sequence lengths). Moreover, we show that these benefits scale substantially along with sequence length, making them indispensable for practitioners aiming to model the full complexity of hospital data. As machine learning continues to grow in healthcare, tight collaborations between hospitals and technology manufactures are thus essential to allow for greater compute resources to input higher-quality data into machine learning models.
Resources
- Dell XE9680 Technical Guide: https://www.delltechnologies.com/asset/en-ca/products/servers/technical-support/poweredge-xe9680-technical-guide.pdf
- Dell R750xa Technical Guide: https://i.dell.com/sites/csdocuments/Product_Docs/en/poweredge-r750xa-technical-guide.pdf
- Dell XE8545 Technical Guide: https://i.dell.com/sites/csdocuments/product_docs/en/poweredge-xe8545-technical-guide.pdf
Authors:
Jonathan Huang, MD/PhD Candidate, Research & Development, Northwestern Medicine
Matthew Wittbrodt, Solutions Architect, Research & Development, Northwestern Medicine
Alex Heller, Director, Research & Development, Northwestern Medicine
Mozziyar Etemadi, Clinical Director, Advanced Technologies, Northwestern Medicine
Bhavesh Patel, Sr. Distinguished Engineer, Dell Technologies
Bala Chandrasekaran, Technical Staff, Dell Technologies
Frank Han, Senior Principal Engineer, Dell Technologies



